TEST_004 The user receives an interpretative distribution representation of possible ICD categories represented in the pixels of the image
Test type
System
Linked activities
- MDS-103
Result
- Passed
- Failed
Description
Tests were carried out for the algorithm that generates a distribution representation of possible ICD categories, to verify its ability to extract ICD-11 category features from the image pixels with high performance.
Objective
The goal is to prove that the output distribution of ICD categories matches the ground truth ICD category.
Acceptance criteria
The sum of confidence values must equal 100%, and we set a minimum requirement of:
- 55% for the top 1
- 70% for the top 3
- 80% for the top 5
For the indicators, we expect a minimum AUC value of 0.80.
Materials & methods
Dataset
The dataset was created by combining images from the following datasets:
- Dermatology image datasets:
- ACNE04: Facial acne images of different severities (mild, moderate, severe).
- ASAN: clinical images of 12 types of skin diseases.
- Danderm: An online dermatology atlas focused on common skin diseases.
- Derm7pt: dermoscopic images of melanoma and nevi cases.
- Dermatology Atlas: a collection of clinical images of subjects of different ages and skin tones,
- DermIS: contains information on skin diseases, illustrated by photographic images, along with descriptions, therapeutic measures, and skin care suggestions.
- DermnetNZ: one of the largest and most diverse collections of clinical, dermoscopic, and histology images of various skin diseases.
- Hallym: clinical images of basal cell carcinoma.
- Hellenic Dermatology Atlas: a dermatology atlas with clinical images of a large set of skin diseases
- ISIC: a collection of clinical and dermoscopic skin lesion datasets from across the world, such as ISIC Challenges datasets, HAM10000, and BCN20000.
- PAD-UFES-20: clinical close-up images of six different types of skin lesions: basal and squamous cell carcinoma, actinic keratosis, seborrheic keratosis, Bowen's disease, melanoma, and nevus.
- PH2: dermoscopic images of melanoma and nevi.
- Severance (validation): this subset comprises benign and malignant tumours.
- SD-260: A collection of images of 260 skin diseases, captured with digital cameras and mobile phones.
- SNU: a subset of images representing patches of skin lesions of 134 diseases (5 malignant and 129 non-malignant).
- Image datasets related to skin structure: these were included to ensure that healthy skin images were also present during training and validation and to prevent the models from overfitting.
- 11kHands: a collection of images of the hands of 190 subjects of varying ages.
- Figaro1k: a database with unconstrained view images containing various hair textures of male and female subjects.
- Foot Ulcer Segmentation (FUSeg): natural images of wounds.
- Hand Gesture Recognition (HGR): gestures from Polish Sign Language, American Sign Language, and special characters. HGR contains images of hands and arms.
This resulted in a dataset with a total size of 241,298 RGB images of nearly 1,000 different classes. As each source followed a different taxonomy to name the ICD class observed in the images, we conducted a thorough revision of all the terms observed in the dataset to arrive at a curated list of usable ICD categories. This revision was done in collaboration with an expert dermatologist.
Additional annotation
We conducted a rigorous curation process involving data enrichment, diversification, and quality assurance procedures. In summary, this process involved:
- Manually crop regions of interest, subsequently utilized as additional training images.
- Manually tag images as clinical, dermoscopic, histology, skin (but not dermatology), and out of domain. The histology and out-of-domain images are discarded from the dataset.
- Manually tag the visible body parts that can be seen in an image.
Diversity and representation of age, sex, and skin tone
For the representation of skin tone, we analyzed the distribution of Fitzpatrick skin types across the entire dataset:
| Fitzpatrick skin type | Number of ICD categories | % images |
|---|---|---|
| FST-1 | 840 | 27.02% |
| FST-2 | 1042 | 36.87% |
| FST-3 | 883 | 24.04% |
| FST-4 | 732 | 7.89% |
| FST-5 | 543 | 3.05% |
| FST-6 | 309 | 1.13% |
In terms of age and sex representation, it was not possible to retrace the data at the same scale as it was not originally available in many of the original datasets. Moreover, due to the characteristics of a high percentage of the images (i.e. dermoscopy or clinical, close-up images), it was not possible to determine the age and sex of the subject just from the image. However, we analyzed the distribution of age and sex in smaller subsets (3,614 and 2,338 images, respectively).
| Sex | % images |
|---|---|
| Male | 51.47 |
| Female | 48.53 |
| Age group | % images |
|---|---|
| 0-25 years | 2.82 |
| 25-50 years | 17.96 |
| 50-75 years | 59.15 |
| 75+ years | 20.06 |
Despite being at a lower scale, these analyses reveal that all age groups, sexes, and skin tones are represented in the dataset.
It is also crucial to understand that some ICD categories and some clinical signs have different population incidence. This means that it is inherently impossible to achieve an evenly distributed dataset across all ages, sexes, and phototypes. For instance, pediatric issues affect children disproportionately, which makes it impossible to achieve an equal representation of pediatric issues across all ages. Moreover, the nature of a large percentage of the images (clinical close-ups or dermoscopy images) and the training methodology used (lesion cropping augmentations) help reduce the model's bias towards sexual and age-related macroscopic features derived from the current imbalanced yet diverse dataset.
Data stratification
Due to the multi-source nature of the dataset, it was possible to apply a patient-wise stratification of the dataset for some of the sources, while the remaining ones were split into train/validation/test class-wise (to ensure the representation of each class is equal in every subset).
Thanks to the volume of the dataset, it was possible to split the dataset into a training, validation, and test set. In addition, some sources we included exclusively in the test set to obtain a good understanding of model performance on unseen data.
| Patient-wise train/val/test split | Class-wise train/val/test split |
|---|---|
| 11kHands, ACNE04, Derm7pt, Figaro1k, FUSeg, HGR, ISIC, PAD-UFES-20, PH2. | ASAN, Danderm, Dermatology Atlas, DermIS, DermnetNZ, Hallym, Hellenic Dermatology Atlas, SD-260, SNU, Severance. |
Data augmentation
Additionally, we employed dataset-specific data augmentation techniques. For instance, for dermatoscopic images, we applied various types of rotations to ensure rotation invariance, which is crucial for this image category. However, such rotation augmentation was not used for clinical images since certain elements, like faces, consistently maintain their position.
Furthermore, we introduced a variety of pixel transformations (colour jittering, histogram equalization, random noise, Gaussian blur, motion blur, etc.) that are applied to all images to enhance the robustness and diversity of the training data. Thanks to these pixel transformations, models can become color-agnostic and focus on texture features, which can help overcome some unwanted biases such as skin tone.
No augmentations were applied to the validation and test data.
Model architecture and training setup
In line with the requirements, we implemented the vision transformer (ViT) on our dataset. Regarding the training and learning policy, we used a one-cycle learning rate policy to enable super-convergence (i.e. enabling a network to learn a task in less time), as described in “Super-Convergence: Very Fast Training of Neural Networks Using Large Learning Rates” (Smith and Topin, 2019).
Results
The algorithm accurately provides a proportional confidence distribution, thanks to its final layer being a softmax layer that ensures the sum of all values equals 100%.
The algorithm achieves compelling top 1, 3, and 5 metrics, with test scores of 74%, 86%, and 90%, respectively.
Moreover, the AUC for malignancy quantification is exceptionally high at 0.96. The dermatological condition indicator achieves an outstanding AUC of 0.99, while the urgent and high-priority referral indicators reach 0.97 and 0.93, respectively.
Bias analysis
Model performance and skin tone
In order to assess the performance gap across populations, the Diverse Dermatology Images (DDI) dataset was proposed by Daneshjou et al. (Disparities in Dermatology AI Performance on a Diverse, Curated Clinical Image Set). This dataset includes malignancy and skin type metadata (divided into 3 groups, i.e. 1-2, 3-4, and 5-6). The authors recorded significant performance drops when using other devices on this dataset. We decided to conduct a similar analysis and evaluated the malignancy prediction performance. Our motivation behind conducting a bias analysis on malignancy estimation over other tasks performed by the device is to provide results that can be directly compared to others obtained with different devices and machine learning algorithms (as in the work by Daneshjou et al.).
The results are almost identical to those reported by the authors. However, after a closer inspection of the DDI dataset (656 images), we detected some critical issues with the dataset that could potentially limit its power. The first was the incorrect framing of the object of interest, followed by poor overall image quality. To improve the quality of the analysis, we manually cropped the 656 images and ran the model on the cropped images.
| Fitzpatrick skin type | AUC (full image) | AUC (cropped lesion) |
|---|---|---|
| 1-2 | 0.6255 | 0.6903 |
| 3-4 | 0.6537 | 0.8365 |
| 5-6 | 0.6417 | 0.6724 |
| Overall | 0.6510 | 0.7627 |
After cropping the images, performance improved for all skin type groups, but the most benefitted group was 3-4. These results agree with already published evidence of models performing better on light and medium skin tones, but they must be taken with caution given the evident limitations of the DDI dataset (sample size and image quality). Skin tone-related features are deeply entangled with quality-related features or non-skin features: in other words, it is difficult to determine if such performance drop for groups 1-2 and 5-6 is due solely to skin tone underrepresentation or other artifacts affecting those specific images.
To get a better understanding of model performance across skin tones, we finally computed the same metric on the entire main dataset, and the results were notably superior.
| Fitzpatrick skin type | AUC (Malignancy) | AUC (Urgent referral) | AUC (High-priority referral) |
|---|---|---|---|
| FST-1 | 0.9309 | 0.9844 | 0.9221 |
| FST-2 | 0.9396 | 0.9720 | 0.9259 |
| FST-3 | 0.9375 | 0.9718 | 0.9378 |
| FST-4 | 0.9273 | 0.9514 | 0.9161 |
| FST-5 | 0.9535 | 0.9451 | 0.9296 |
| FST-6 | 0.9409 | 0.9315 | 0.9268 |
Regarding skin image recognition and the other tasks described in other tests (i.e. quantification of intensity, count, and extent of visible clinical signs), there is a generalized lack of skin tone labels in most common skin image datasets and atlases, which made it difficult to conduct similar analyses. The characterization of skin type in the main dataset enables us to conduct similar experiments in the future and provide results for other processors.
Visual bias explainability
Apart from exploring biased performance on the DDI dataset, we have implemented GradCAM for the skin image recognition model. This lets us inspect model activations to an image's visual features at inference time, which is also helpful for the detection of unwanted biases. The gradient-weighted class activation map (GradCAM) method is a popular visualization technique useful for understanding how the model has been driven to make a classification decision. It is class-specific, providing separate visualizations for each class in the dataset.
Protocol deviations
There were no deviations from the initial protocol.
Conclusions
The algorithm generates a distribution representation of possible ICD categories with a high performance. This ensures the quality of the data, offering healthcare practitioners the best information to support their clinical assessments.
Test checklist
The following checklist verifies the completion of the goals and metrics specified in the requirement REQ_004.
Requirement verification
- The output contains the ICD code and confidence value
- The sum of the confidence values add up to 100%
- top-5 accuracy > 80%
- top-3 accuracy > 70%
- top-1 accuracy > 55%
- AUC > 0.8 in malignancy quantification
- AUC > 0.8 in the quantification of the presence of a dermatological condition
- AUC > 0.8 in the quantification of referral urgency
- AUC > 0.8 in the quantification of referral priority
Evidence
To verify the algorithm output contains the ICD code and confidence value we run the algorithm and provide a screenshot with the output:

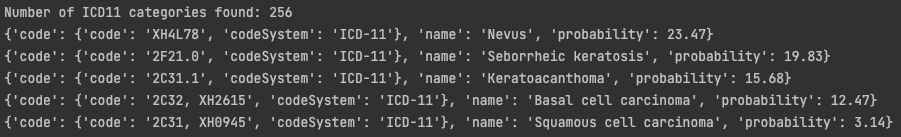
To prove confidence values add up to 100%, we sum the output of the past test and provide a screenshot of the result:

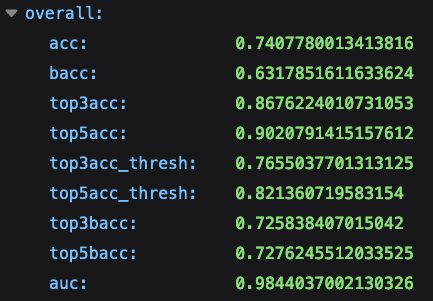
Evidence of top N accuracy values can be found in the output JSON generated by our experiments. The JSON is evaluated in an independent set of N images containing all the ICD-11 categories learned by the algorithm.
The following metrics can be found in the JSON file:
The output of the binary indexes that are calculated based on ICD-11 categories and their confidence can also be checked by running the algorithm for several images and checking the output:
Information about each indicator's performance is provided in JSON format. The malignancy indicator achieves an AUC of 0.96 ± 0.07, the dermatological condition indicator scores 0.99, and the referral indicators attain values of 0.97 and 0.93.
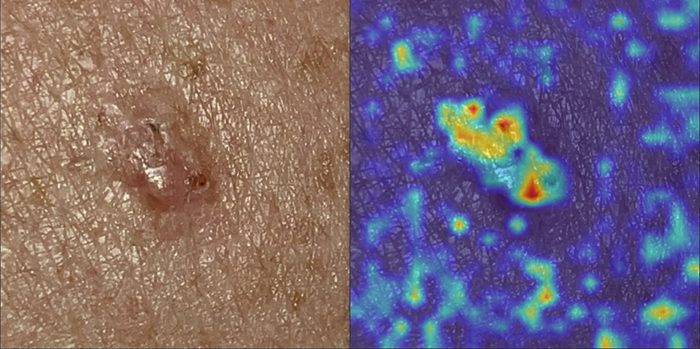
This is an example of explainable skin disease recognition with GradCAM. In this case, the test image corresponds to a basal cell carcinoma, and the heat map on the right side shows the activated areas for that class:
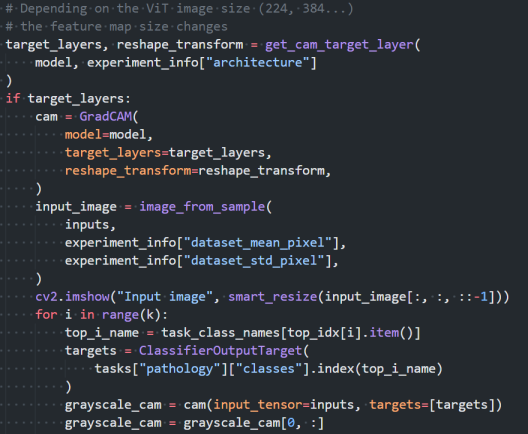
GradCAM is used internally to analyse model performance:
Signature meaning
The signatures for the approval process of this document can be found in the verified commits at the repository for the QMS. As a reference, the team members who are expected to participate in this document and their roles in the approval process, as defined in Annex I Responsibility Matrix of the GP-001, are:
- Tester: JD-017, JD-009, JD-004
- Approver: JD-005